Implementing PubMed 200k RCT Paper with TensorFlow
As an AI enthusiast and a passionate learner, I recently embarked on the exciting adventure of implementing the PubMed 200k RCT paper using TensorFlow. In this blog post, I’ll share my experiences, challenges, and insights gained during this fascinating endeavor.

Understanding PubMed 200k RCT
What is PubMed 200k RCT?
PubMed 200k RCT is a remarkable dataset designed for sequential sentence classification in medical abstracts. Created by Franck Dernoncourt and Ji Young Lee, this dataset is based on PubMed, a treasure trove of scientific literature. Let’s dive into the specifics:
- Dataset Size: Approximately 200,000 abstracts from randomized controlled trials (RCTs), totaling a whopping 2.3 million sentences12.
- Sentence Labels: Each sentence in an abstract is meticulously labeled with its role, falling into one of the following classes: background, objective, method, result, or conclusion.
- Purpose: The release of this dataset serves two critical purposes:
- Algorithm Development: Most existing datasets for sequential short-text classification are small. By introducing PubMed 200k RCT, we aim to foster the development of more accurate algorithms for this task.
- Efficient Literature Skimming: Researchers often need efficient tools to navigate through extensive literature. Automatically classifying each sentence in an abstract can significantly enhance researchers’ productivity, especially in fields where abstracts tend to be lengthy, such as medicine.
The Implementation Journey
1. Data Preparation
Before diving into model architecture, I meticulously prepared the data:
- Data Collection: I retrieved the PubMed 200k RCT dataset, ensuring I had access to the abstracts and their corresponding sentence labels.
- Preprocessing: Cleaning and tokenizing the text were essential steps. I removed noise, handled special characters, and split sentences into tokens.
- Model Input overview:

- Model Output Overview:

2. Model Architecture
For the heart of our implementation, I chose TensorFlow as my deep learning framework. Here’s how I structured the model:
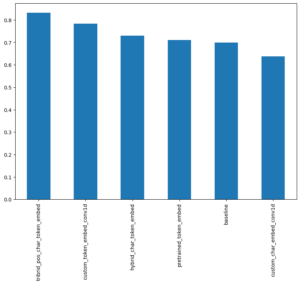
- Base Model:TF-IDF Multinomial Naive Bayes as recommended by [Scikit-Learn’s machine learning map](https://scikit-learn.org/stable/tutorial/machine_learning_map/index.html).
-
Model 1: Conv1D with token embeddings
- Model 2: Feature extraction with pretrained token embeddings
- Model 3: Conv1D with character embeddings
- Model 4: Combining pretrained token embeddings + character embeddings (hybrid embedding layer)
-
Model 5: Transfer Learning with pretrained token embeddings + character embeddings + positional embeddings
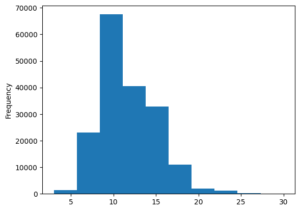
Most of the abstracts are around 7 to 15 sentences in length.
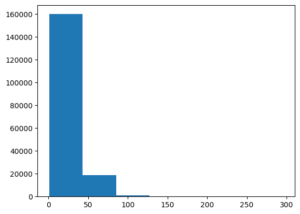
Vast majority of sentences are between 0 and 50 tokens in length
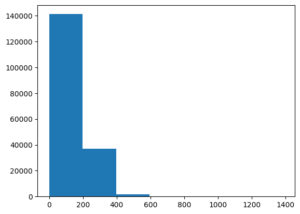
Sequences are between 0 and 200 characters long.
Evaluation Metrics
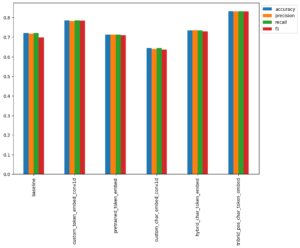
Conclusion
Implementing PubMed 200k RCT with TensorFlow was both challenging and rewarding. I gained a deeper understanding of deep learning, natural language processing, and the importance of large, high-quality datasets. As I continue my AI journey, I’m excited to explore more groundbreaking research and contribute to the ever-evolving field. Github Project link.